The phylogenetic history (how one species relates to another genetically) of madtom catfishes has been under investigation for decades with the first hypothesis published in 1969 and the most recent in 2017. Over the 50+ years since the first hypothesis, there has been several new discoveries and descriptions of madtom catfishes, hypotheses of distinct populations within a species harboring cryptic diversity (genetically distinct species that are indistinguishable morphologically from known species), and relationships between species that have remained unresolved. Therefore, my project aims to resolve these relationships and attempt to tease apart the cryptic diversity within this genus by collecting fresh tissue samples from all 29 known species, with additional samples from unique geographic locations for species hypothesized to be cryptic . DNA will be sequenced using restriction site-associated DNA sequencing (RADseq) methodology to reconstruct novel phylogenetic hypotheses with RAD loci. This research will provide insight into the evolution and diversity of an understudied group of organisms, with many of the species within the genus under federal or state protection.
Presentations
Joint Meeting for Ichthyologist and Herpetologist, 2022
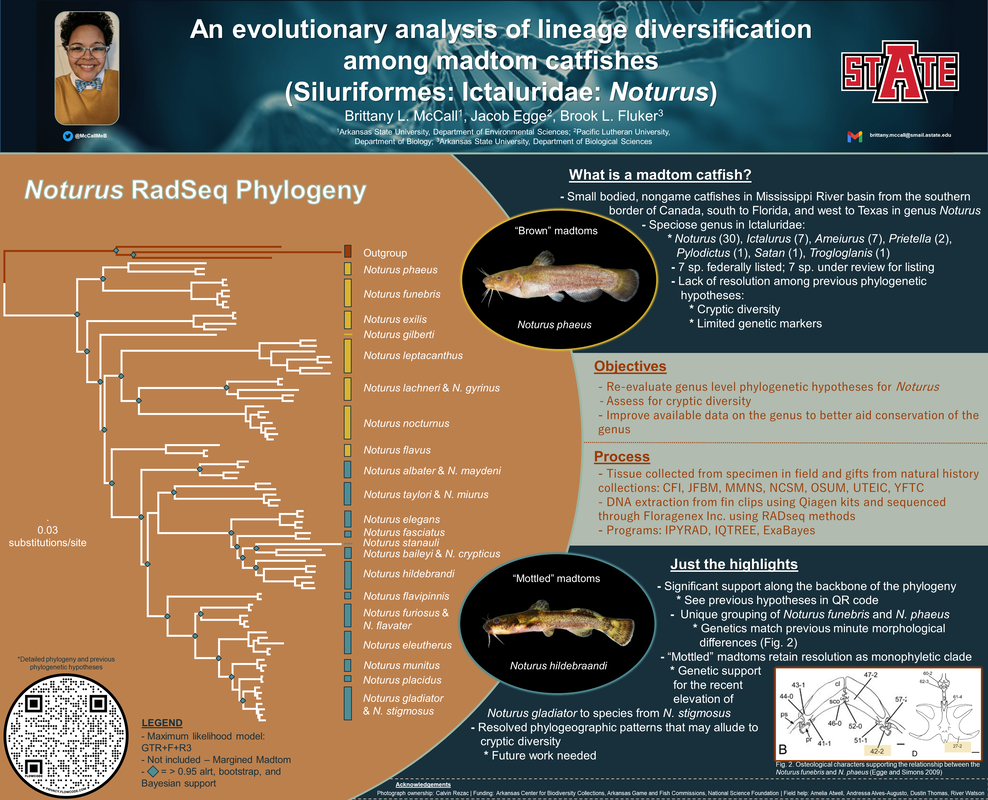
Below is the conference poster for the JMIH 2022 conference with preliminary data. The project at this time featured a maximum likelihood and bayesian inference of RADseq data for all species, except Noturus insignis and the Broadtail Madtom. Preliminarily, we show significant support for deeper relationships that has not been found previously, the possibility of a independent lineage sister to all Noturus, novel relationships, and corroborated patterns of intraspecific diversity that may elude to cryptic diversity. Additional data to come a comparison to the whole genome for Noturus placidus, divergence and species tree analyses, and the inclusion of the N. insignis group.
Below is the conference poster for the JMIH 2022 conference with preliminary data. The project at this time featured a maximum likelihood and bayesian inference of RADseq data for all species, except Noturus insignis and the Broadtail Madtom. Preliminarily, we show significant support for deeper relationships that has not been found previously, the possibility of a independent lineage sister to all Noturus, novel relationships, and corroborated patterns of intraspecific diversity that may elude to cryptic diversity. Additional data to come a comparison to the whole genome for Noturus placidus, divergence and species tree analyses, and the inclusion of the N. insignis group.
Wonders of the Creeks
Just a few of my favorite finds while collecting tissue samples for this project.